How Many Domains of Life?
September 13, 2011
Classification is an interesting problem. There's a box of
miscellaneous bolts in my workshop, commonly called a "hell box" by
machinists, where I often glean a piece or two for a project. If I were to classify the contents of this box, what characteristics of these bolts would I use?
As someone who dabbles in
computer science, I would use the classification scheme that makes the most sense from a search standpoint. In this case, it would be material type (
steel,
brass,
nylon, etc.), then size (no. 2, 4, 6, 8, etc.), then
thread pitch (20, 32, 40, etc). For me, material type is the most important characteristic, and this specifies my three "domains" for bolts.
Biologists have the same problem in the classification of
life, and the most fundamental question is how many domains of life are there? The first attempt at classification recognized just two
domains. These were the
Linnaean "super-kingdoms" of "
prokaryotes" and "
eukaryotes." Eukaryote cells (
animals,
plants and
fungi) have a
nucleus, and prokaryote cells (
bacteria) do not.
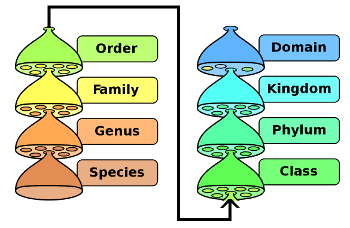
Biological classification.
Domain is the top level of species classification.
(Image by Peter Halasz (modified) via Wikimedia Commons))
A little more than thirty years ago, another domain was added; namely, the
archaea, which I wrote about in a
previous article (Archaea, November 30, 2007).[1-4] This domain was proposed by
Carl Woese on the basis of something more fundamental than looking through a
microscope for a cell nucleus. Woese proposed a segmentation according to
ribosomal RNA (rRNA).
Woese became interested
methanogens, bacteria that produce
methane
instead of
carbon dioxide in
metabolism. This strange sort of metabolism notwithstanding, these were still classified as bacteria. Woese sequenced the ribosomal RNA from one such species,
M. bryantii, and he found it to be quite unlike that of other bacteria; so different, that he proposed the existence of a third domain of life, the
Archaea.[4]
The figure below shows these three domains, Bacteria, Archaea and
Eucarya, also called Eubacteria, Archaebacteria and Eukaryota.
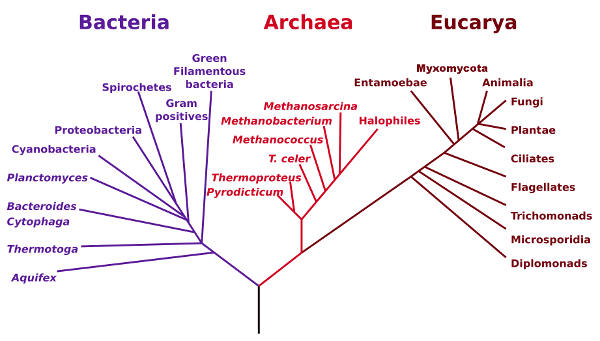
Phylogenetic Tree of Life. (NASA image via Wikimedia Commons).
Of course, getting
scientists in any field to agree on something is difficult. Most follow the three domain system, but some still cling to the two super kingdoms of eukaryotes and prokaryotes. The Archaea are so rare that on a practical level, it doesn't really matter.
There's also the
six-kingdom system of
Eubacteria,
Archaebacteria,
Protista,
Fungi,
Plantae, and
Animalia. This allows a top-level differentiation between plants and animals that's satisfying. What's important depends on whom you ask. As a non-voting, non-biologist, I'm a member of the three domain camp.
But wait! There's more! A recent news focus article in
Nature by
Gwyneth Dickey Zakaib reviews evidence for yet another domain of life.[5] This domain is that of
giant viruses, and there may be others on the sidelines as evidenced by unexplained
marine DNA.
The
modus operandi of
viruses is that they infect
cells and hijack cellular mechanism for their own use. They can't reproduce outside of a host cell. To do what little they do, viruses don't need a very complicated
genome, and they don't need to be large. A small size is actually an advantage.
That's why a virus discovered in 1992 by
Didier Raoult of the
University of the Méditerranée in
Marseille, France, caused such a stir. This
Mimivirus, the first so-called "giant virus," had a total length of 600
nm; that is, it's so large that it's visible under an
optical microscope. Not only that, but its DNA has 1.2 million
base pairs.[5]
Raoult's team discovered another giant virus, named
Marseillevirus, in 2009. Isolated from
amoebae, it has a 368 kilo-base-pair genome that encodes at least 49
proteins. Raoult considers the Mimivirus, Marseillevirus, and their giant virus cousins, to be part of a new domain of life, a fourth domain.
This new domain is called, quite non-poetically,
Nucleocytoplasmic Large DNA Viruses (NCLDVs). Raoult's proposal has met considerable resistance, just as Woese's third domain did years ago. Once again, on a practical level, it doesn't really matter, since the giant viruses are quite rare.
The rarity argument against other domains of life might not hold water (pun intended) considering research into uncategorized DNA sampled from the
oceans. Such DNA has been called the "dark matter of the biological universe" by
Jonathan A. Eisen of the
University of California Davis (Davis, California).[5]
Eisen is an author of a
PLOS One article entitled, "Stalking the Fourth Domain...," that examines these DNA sequences and
metagenomic data.[6] The sequences are far removed from those of the known domains, and thus the mention of a fourth domain in his article's title.
Francisco Rodriguez-Valera a
microbiologist at the
Miguel Hernández University (Alicante, Spain), sums up the consensus in his field this way.
"There is a huge amount of microbial diversity that is unknown. I don't think we need to discover a new domain every ten years to convey to the general public the fact that microbes are important."[5]
References:
- Diana Yates, "Symposium marks 30th anniversary of discovery of third domain of life" (University of Illinois Press Release, October 16, 2007).
- George E. Fox, Linda J. Magrum, William E. Balch, Ralph S. Wolfe and Carl R. Woese, "Classification of Methanogenic Bacteria by 16S Ribosomal RNA Characterization," Proc. Natl. Acad. Sci., vol. 74, no. 10 (October 1, 1977), pp.4537-4541.
- Carl R. Woese and George E. Fox, "Phylogenetic Structure of the Prokaryotic Domain: The Primary Kingdoms," Proc. Natl. Acad. Sci. vol. 74, no. 11 (November 1, 1977), pp. 5088-5090.
- Carl R. Woese, Otto Kandler and Mark L. Wheelis, "Towards a natural system of organisms: proposal for the domains Archaea, Bacteria, and Eucarya," Proc. Natl, Acad. Sci., vol. 87, no. 12 (June 1, 1990), pp. 4576-9 .
- Gwyneth Dickey Zakaib, "The challenge of microbial diversity: Out on a limb," vol. 476, no. 7358 (August 4, 2011) pp. 5-120.
- Dongying Wu, Martin Wu, Aaron Halpern, Douglas B. Rusch, Shibu Yooseph, Marvin Frazier, J. Craig Venter and Jonathan A. Eisen, "Stalking the Fourth Domain in Metagenomic Data: Searching for, Discovering, and Interpreting Novel, Deep Branches in Marker Gene Phylogenetic Trees," PLoS ONE, vol. 6, no. 3 (2011), Document No. e18011.
Permanent Link to this article
Linked Keywords: Classification; randomness; bolt; machinist; computer science; steel; brass; nylon; thread pitch; biologist; organism; life; domain; Linnaean; prokaryotes; eukaryotes; animal; plant; fungus; nucleus; bacteria; Wikimedia Commons; archaea; Carl Woese; microscope; ribosomal RNA; methanogen; methane; carbon dioxide; metabolism; methanobacterium; M. bryantii; Archaea; Eucarya; Phylogenetic Tree of Life; NASA; scientist; six-kingdom system; Eubacteria; Archaebacteria; Protista; Fungi; Plantae; Animalia; Ronco; But wait! There's more!; Nature; Gwyneth Dickey Zakaib; giant viruses; marine; DNA; modus operandi; viruses; cell; genome; Didier Raoult; University of the Méditerranée; Marseille, France; Mimivirus; nanometer; nm; optical microscope; base pair; Marseillevirus; amoeba; protein; Nucleocytoplasmic Large DNA Viruses; ocean; Jonathan A. Eisen; University of California Davis (Davis, California); PLOS One; metagenomic; Francisco Rodriguez-Valera; microbiologist; Miguel Hernández University (Alicante, Spain).